Image-Centric Integrated Data Model of Medical Information by Diseases: Two Case Studies for AMI and Ischemic Stroke
Article information
Abstract
In the medical fields, many efforts have been made to develop and improve Hospital Information System (HIS) including Electronic Medical Record (EMR), Order Communication System (OCS), and Picture Archiving and Communication System (PACS). However, materials generated and used in medical fields have various types and forms. The current HISs separately store and manage them by different systems, even though they relate to each other and contain redundant data. These systems are not helpful particularly in emergency where medical experts cannot check all of clinical materials in the golden time. Therefore, in this paper, we propose a process to build an integrated data model for medical information currently stored in various HISs. The proposed data model integrates vast information by focusing on medical images since they are most important materials for the diagnosis and treatment. Moreover, the model is disease-specific to consider that medical information and clinical materials including images are different by diseases. Two case studies show the feasibility and the usefulness of our proposed data model by building models about two diseases, acute myocardial infarction (AMI) and ischemic stroke.
1. Introduction
Medical systems including Hospital Information System (HIS) are constantly evolving for digitalizing and efficiently managing medical information. Nowadays most of medical institutions like hospitals naturally use HIS. Recently, many studies [1–10] have tried to efficiently store, transfer, and manage clinical materials. However, HIS consists of several systems such as Order Communication System (OCS), Electronic Medical Record (EMR), and Picture Archiving and Communication System (PACS) and they generate clinical materials of different types. Those systems tend to separately and redundantly include one patient’s information. In other words, since information about a patient’s condition is contained in various materials, medical experts should read all of charts or reports and interpret all of images or videos before diagnosing. Therefore, systems should be able to provide and manage essential data required in emergency so that medical experts can make a quick decision.
Therefore, one of the primary issues in HIS is how to effectively manage and provide a patient’s information for medical experts, particularly in urgent situations. Some institutions have presented schemes for describing medical and health data [12–14]. However, even though these standards are able to represent and integrate general medical data, it is hard to describe the detailed data and semantics dependent on a specific disease by using them.
In this paper, we propose a new data model which integrates medical information by diseases. The model can represent common medical data as well as disease-specific data. Besides, the data model will support the HISs in order to provide essential data for medical experts who should immediately make a decision about the diagnosis and treatment.
This paper is organized as follows: Section 2 discusses the existing works for medical systems and medical information management. In Section 3, we propose a process to build an integrated data model for a specific disease. Section 4 presents the details of the proposed data model through case studies. Section 5 concludes this paper.
2. Related Works
After HISs have become common, many efforts are ongoing to efficiently represent medical information in EMR [1,2] and to efficiently manage them [3,4]. Some previous works presented analysis or processing methods [5–8] and transfer or exchange systems [9,10] for medical images. However, since they have focused on a specific type of HISs or clinical materials, they did not consider the diversity and semantic relations of medical data in multi-systems. Recently, a few studies have tried to utilize various types of medical data for medical image retrieval using semantic relations with radiology reports [11] and for diagnosis using textural and morphological data [15].
Moreover, many efforts have been taken for the efficient modeling and management of personal health information including medical data. Some of them have focused on a specific type among various clinical materials. [16–18] presented ontology models to analyze and represent semantic data implicitly contained in medical images or videos. However, the scope of those models is limited to just one type of clinical materials and medical systems (PACS).
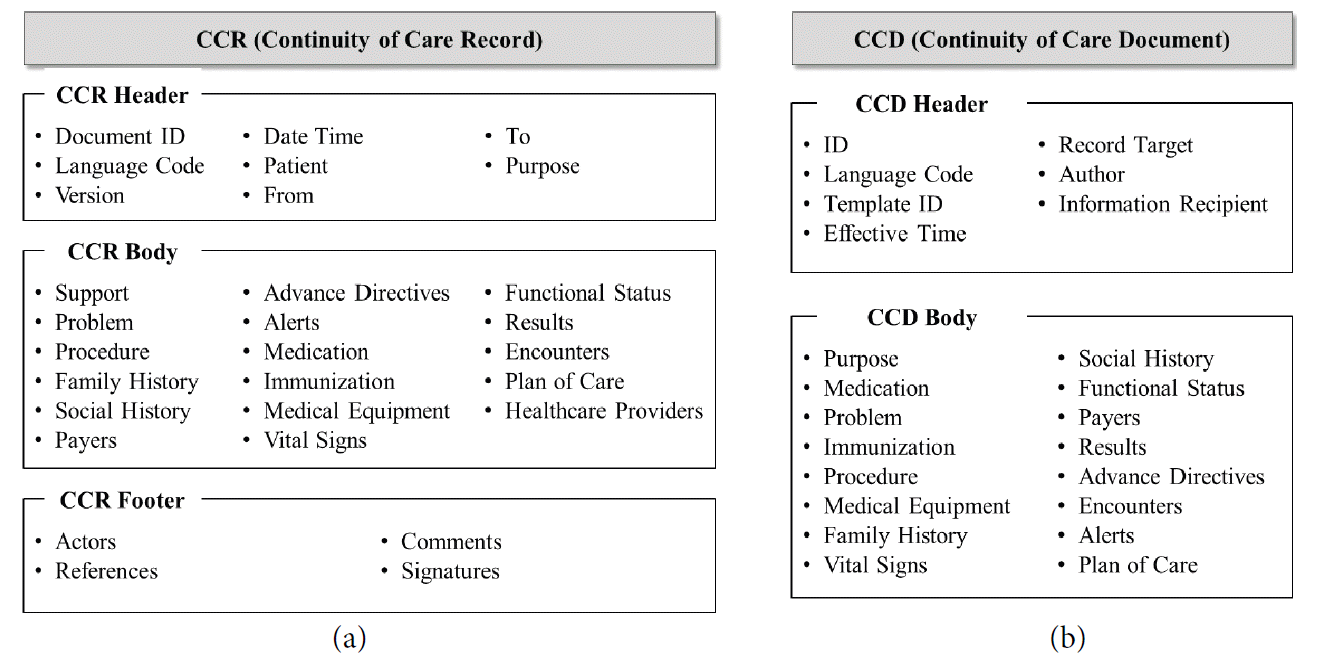
As the specifications for personal health data, CCR, CDA, and CCD are representative standards presented by international institutions, American Society for Testing and Materials (ASTM) and Health Level 7 (HL7). Continuity of Care Record (CCR) [12] is a health record standard specification to contain all of a patient’s relevant medical history. Fig. 1(a) shows the overview of CCR schema. Clinical Document Architecture (CDA) [13] is a standard to specify the structure and semantics of clinical documents. Continuity of Care Document (CCD) [14] shown in Fig. 1(b) is an XML-based markup standard for specifying a summary of a patient’s clinical document. These standards can describe general medical and health information. However, they cannot represent particular data dependent on a specific disease.
3. Integrated Data Model of Medical Information
We suggest a new data model which has the following characteristics or strengths in order to manage vast medical data as a single record. With the data model, a HIS can efficiently manage a patient’s information and quickly provide core data which is necessary for a decision about the diagnosis and treatment to medical experts.
Data integration: the current HISs manage a patient’s information with diverse materials such as reports (documents), charts, and images/videos. However, it is not easy for medical experts (physicians) to check those materials one by one, particularly in emergency cases. Thus, the proposed model integrates medical data stored in various materials into a single record.
Image-centric: among various clinical materials, the medical images or videos which are generated by medical examinations like magnetic resonance imaging (MRI) and computed tomography (CT) play key roles in diagnosing and treating diseases. Besides, they explicitly and implicitly contain core information about a patient’s conditions. In order to analyze correlations between clinical materials and integrate their data, the proposed data model focuses on medical images/videos.
Disease-specificity: medical information is not identical for all of diseases, since types of medical data, clinical materials, and examinations for the diagnosis and treatment of each disease are different. Even in the current hospitals, medical departments have used charts or reports of their own forms. Therefore, a data model should reflect the disease-specificity.
Fig. 2 shows a process to build an integrated data model for a specific disease. The process can define a specification of data model by analyzing clinical materials and the anatomy, centered at medical images. The following section will explain the details about each step through two case studies.
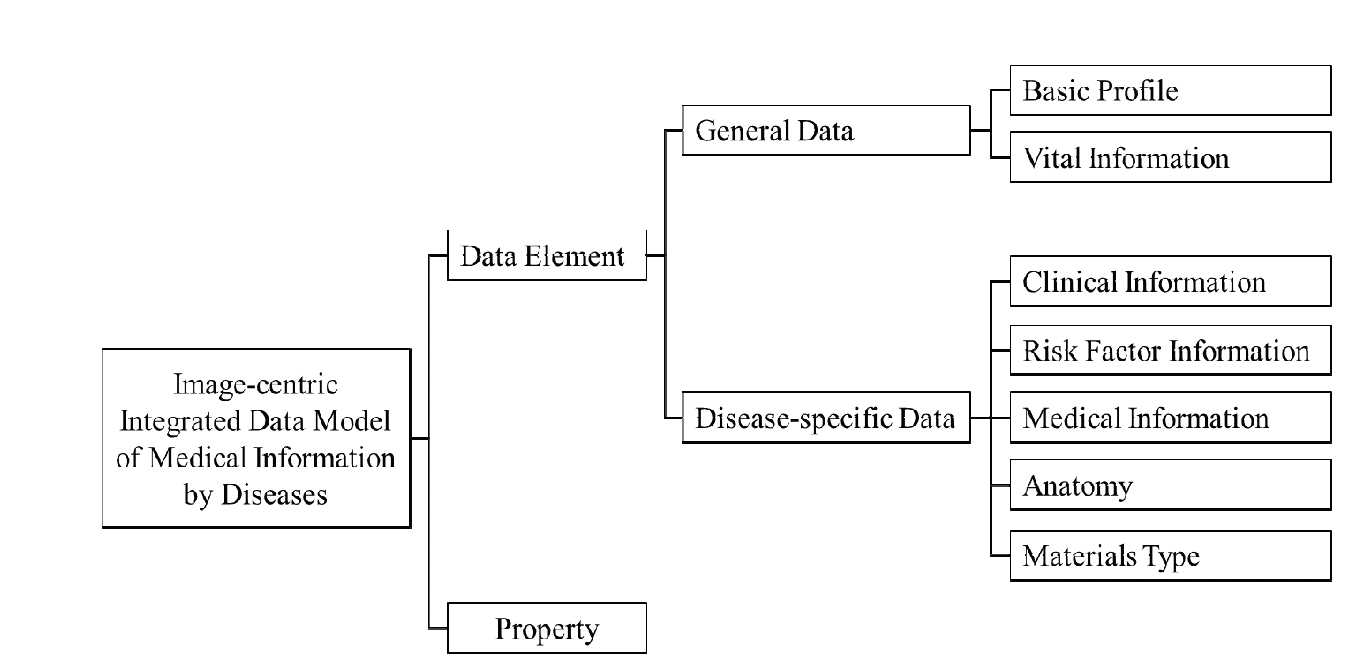
We define a data model as shown in Fig. 3 by applying this process to two diseases. The data model is described as an ontology in order to richly represent relations of integrated data. It consists of two parts; data element and property. The ‘Data Element’ part contains medical data extracted from clinical materials and is organized with hierarchical classes in an ontology. This part is composed of two sub-parts. ‘General Data’ is common to all diseases, and ‘Disease-specific Data’ depend on diseases. The ‘Property’ part includes attributes and relations of data elements. The details of the data model in Fig. 3 will be explained in the next section.
4. Case Study
4.1 Acute Myocardial Infarction
We built a data model for two urgent diseases through the process in Fig. 2. An integrated data model for the first target disease, acute myocardial infarction (AMI), is defined by the following procedures.
Selecting a target disease: AMI occurs as coronary arteries become blocked or narrowed due to a buildup of plaque. When a patient who suspects AMI comes to an emergency room, medical experts should diagnose and treat him/her within 90 minutes at most by checking his/her medical information. Therefore, AMI is a proper case which needs our model.
Selecting core medical image modalities: in a case of AMI, four image/video modalities are essentially used for the diagnosis and treatment and Fig. 4 shows samples. Two modalities are coronary angiography (CAG) and echocardiogram which generate materials of a video type. Electrocardiogram (EKG) produces an image as the examination result. The final modality is a coronary arteriogram which is an image describing a summary of a patient’s condition.
Selecting clinical materials: we chose eight materials including four types of images/videos and four types of reports (documents); CAG, echocardiogram, EKG, coronary arteriogram, CAG report, PTCA and stent deployment report, cardiology lab sheet, and echocardiography laboratory.
Analyzing the anatomy: Since clinical materials notate names of the organ related to a disease, a data model should include anatomical knowledge. AMI is a disease related to the coronary arteries. Then, we analyze the coronary anatomy including right coronary artery (RCA) and left coronary artery (LCA) and their relations.
Analyzing the correlations: the medical images/videos selected in step 2 have relations to the anatomy analyzed in step 4. For example, a CAG video of a specific angiographic view shows certain parts of the whole coronary arteries. These relations will be the basis of an image-based information provision service.
Extracting data: this step analyzes and derives meaningful data explicitly and implicitly contained in eight clinical materials. For instance, a CAG video has explicit data like metadata including date, equipment model and measurement axis. In addition, it has semantic data such as the site of lesion and the degree of severity, which should be interpreted by medical experts.
Classifying data and analyzing their correlations: some of extracted data may be duplicated in multiple materials. Even though some of them are contained in different materials, they may be related to each other. Therefore, after the previous step, we classify extracted data and analyze their relations, regardless of their source materials.
Defining data elements and properties: the final step defines a single record as a data model. The data model is described with classes and properties of ontology schema.
The data model has the structure as shown in Fig. 3. ‘Basic Profile’ of ‘General Data’ shown in Fig. 5 represents personal information of a patient. Fig. 6 shows ‘Vital Information’ which includes fundamental information about medical states.
‘Disease-specific Data’ includes unique data for each disease and consists of the following five sub-categories.
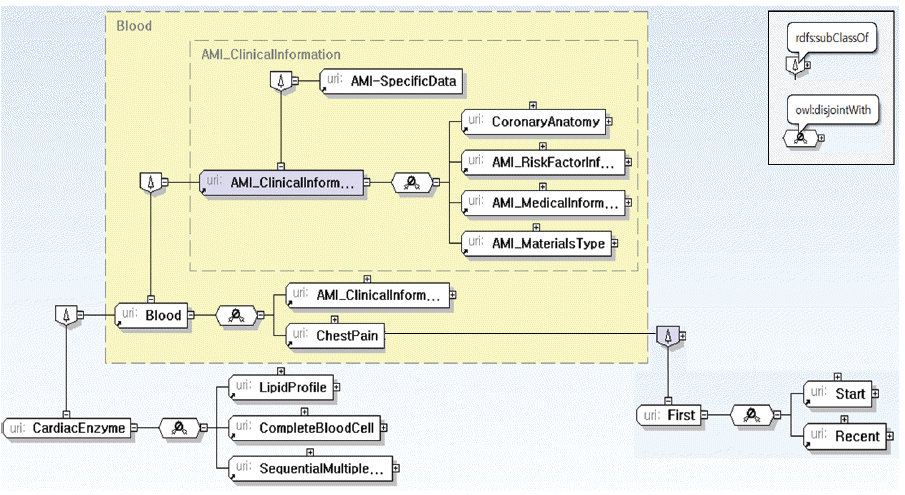
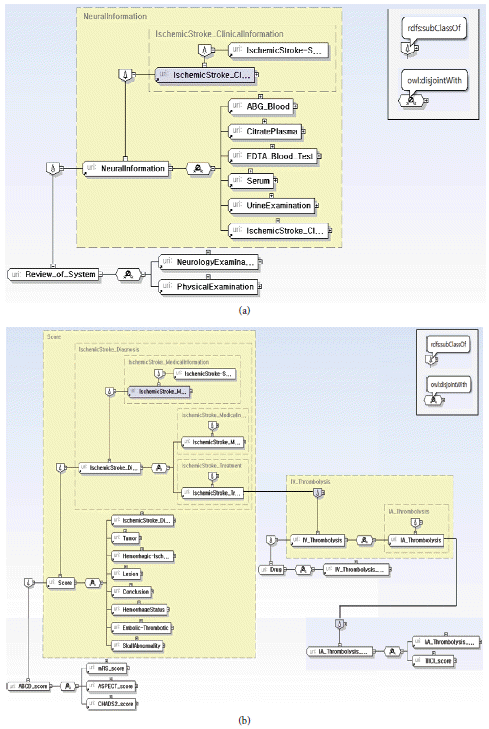
Clinical Information: a patient’s results of various medical examinations except image/video examinations (as shown in Fig. 7).
Materials Type: types of reports, documents, images, and videos (Fig. 8(a)).
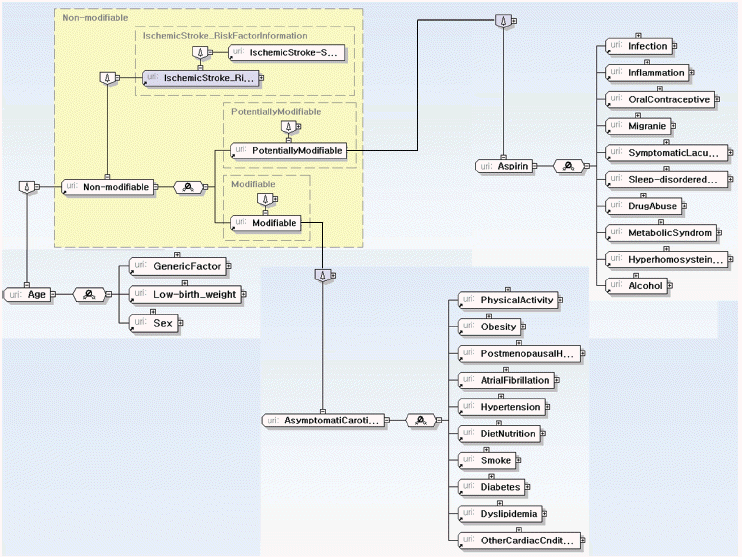
Risk Factor Information: factors, such as habits or environmental conditions, which cause a particular disease (Fig. 8(b)).
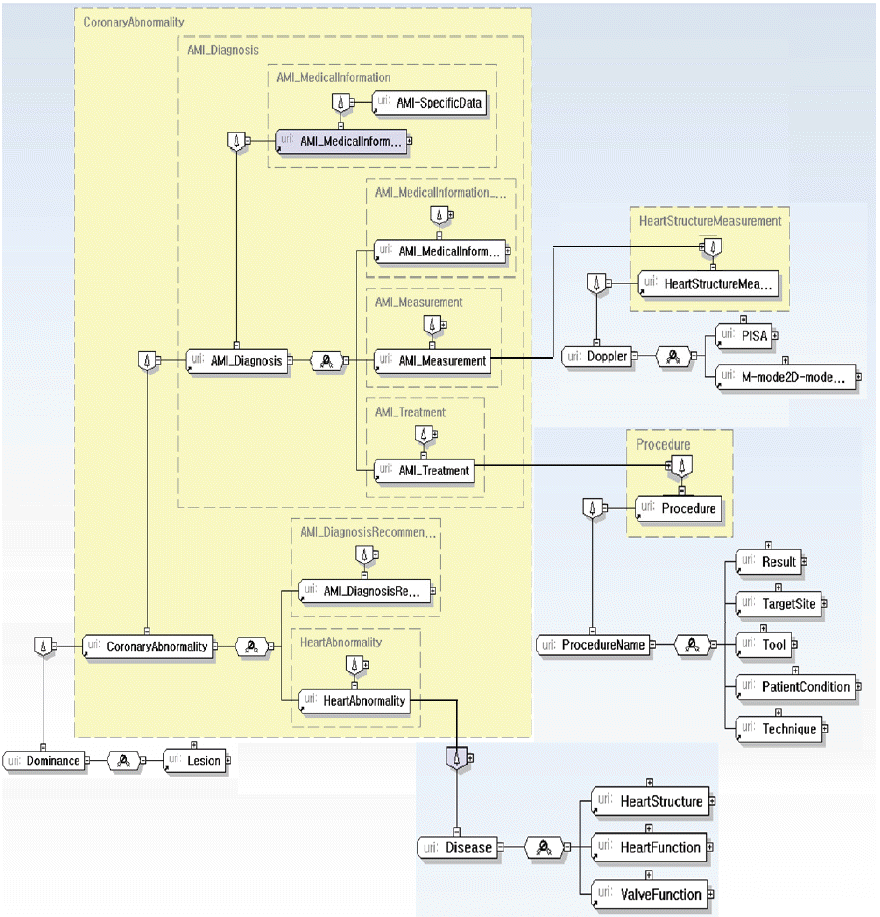
Anatomy: anatomical knowledge for the disease (Fig. 9).
Medical Information: data related to the diagnoses and treatment (Fig. 10).
In addition to data elements described as classes in ontology, the data model represents their attributes and relations as properties. In our case studies, the ‘Property’ is a common part for two diseases, and it has two groups as explained in Table 1. Object property in ontology specifies relations between data elements (classes), and data property means their attributes.
4.2 Ischemic Stroke
The second target disease is ischemic stroke. We briefly explain the procedures to build a data model, since they are similar to that of AMI in the previous sub-section.
Selecting a target disease: ischemic stroke which is one of urgent diseases and occurs when an artery to the brain is blocked.
Selecting core medical image modalities: CT, CT angiography (CTA), MR, transfemoral cerebral angiography (TFCA), and EKG.
Selecting clinical materials: in addition to five images/videos, CT report, CTA report, MR report, TFCA report, and EKG report, clinical laboratory sheet, and neurology exam report.
Analyzing the anatomy: brain anatomy such as brainstem and cerebrovascular supply.
Analyzing the correlations: analyzing relations between the brain anatomy and five images/videos.
Extracting data: analyzing and extracting data from twelve materials.
Classifying data and analyzing their correlations: analyzing redundancy and associations of extracted data.
Defining data elements and properties.
5. Conclusion
The current medical information systems have managed a patient’s information by using different systems like EMR, PACS, and OCS. Therefore, since information even for one patient is stored in heterogeneous materials of various systems, medical experts cannot efficiently check important information, especially in urgent situations. In order to improve the way of managing and providing medical data, we have proposed a new scheme for medical data model. The model can integrate heterogeneous information which is distributed in different systems into a single record. A system can provide essential data, rather than materials or files themselves, for medical experts. Moreover, the model focuses on clinical materials of image of video types, since they include core data about a patient’s condition. In addition, the data model can consider that essential data for the diagnosis and treatment is different by diseases. That is, the proposed data model integrates heterogeneous medical information for a specific disease by focusing on images and videos.
We have applied the proposed building process to two urgent diseases, AMI and ischemic stroke, and then built a data model. The model consists of two parts; the general part represents data common to all of diseases, and the disease-specific part describes data dependent on a specific disease.
Our future works will focus on extending the data model to cover more diseases and implementing an image-based information provision service based on the proposed data model in order to prove its efficiency.
Acknowledgement
This research was supported by the Ministry of Science, ICT and Future Planning (MSIP), Korea, under the Information Technology Research Center (ITRC) support program (IITP-2016-R2718-16-0015) supervised by the IITP (National IT Industry Promotion Agency).
References
Standard specification for Continuity of Care Record (CCR), ASTM E2369-12, 2012.
Biography
Meeyeon Lee

She is a lecture professor of the Department of Electrical and Computer Engineering at Ajou University, Korea. She received Ph.D. degree in Computer Science and Engineering from Ewha Womans University, Korea, in 2012. For 2012–2014, she was a post-doctoral researcher in Ubiquitous Convergence Research Institute (UCRi). She was a research assistant professor in Ajou University, Korea, for 2014–2015. Her research interests include ubiquitous computing, mobile computing, context-awareness, data modeling, bio-medical data modeling, and ontology.
Ye-Seul Park

She is in Master’s course in Department of Electrical and Computer Engineering at Ajou University, Suwon, Korea. She received B.S. degree in Electrical and Computer Engineering from Ajou University in 2015. Her research interests include data modeling for bio-medical images, ontology and embedded software.
Jung-Won Lee

She is an associate professor of the Department of Electrical and Computer Engineering at Ajou University, Korea, since 2006. For 2003–2006, she was the BK professor in Ewha Womans University. She received her Ph.D. degree in Computer Science Engineering from Ewha Womans University, Korea in 2003. She was a researcher of LG Electronics and did an internship in the IBM Almaden Research Center, USA. Her research interests include mobile context awareness, ontology, biomedical data modeling and embedded and software.