1. Introduction
To analyze biomedical literature, some previous approaches have focused only on recognizing named entities (such as proteins), while some recent approaches have emphasized the problem of identifying the interaction between two entities [1–6]. They are interested in extracting binary relations, such as protein-protein interactions and disease-gene associations. However, such binary relations do not provide a deep analysis of biomedical phenomena. Consequently, a bio-event extraction task is required to recognize bio-molecular events that describe a change in the state of the bio-molecular event [7].
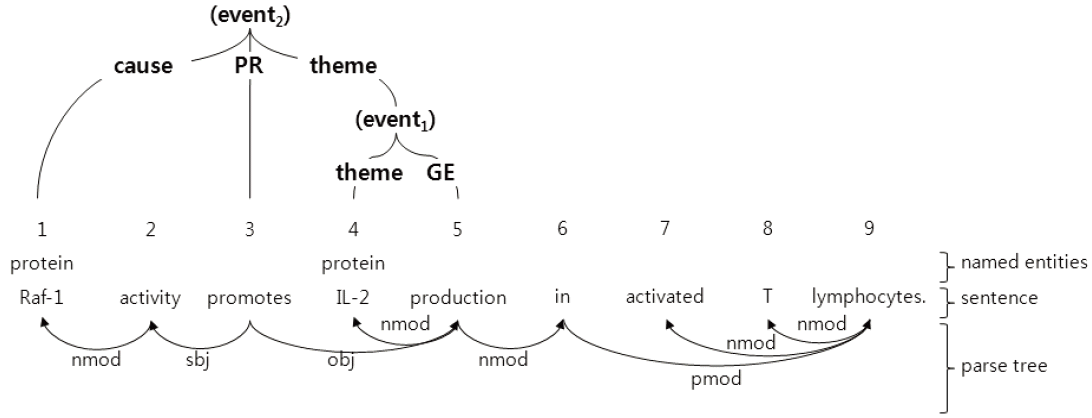
For this task, we tried to identify a set of events where each event consisted of a trigger and its arguments [7]. In the example of Fig. 1, a set {(event1), (event2)} is recognized and the trigger is identified as promotes, and its arguments, such as theme and cause, are also identified. We assumed that the biomedical text was already analyzed with a named entity recognizer, which is a part-of-speech tagger, and a dependency parser, as shown in the lower portion of Fig. 1.
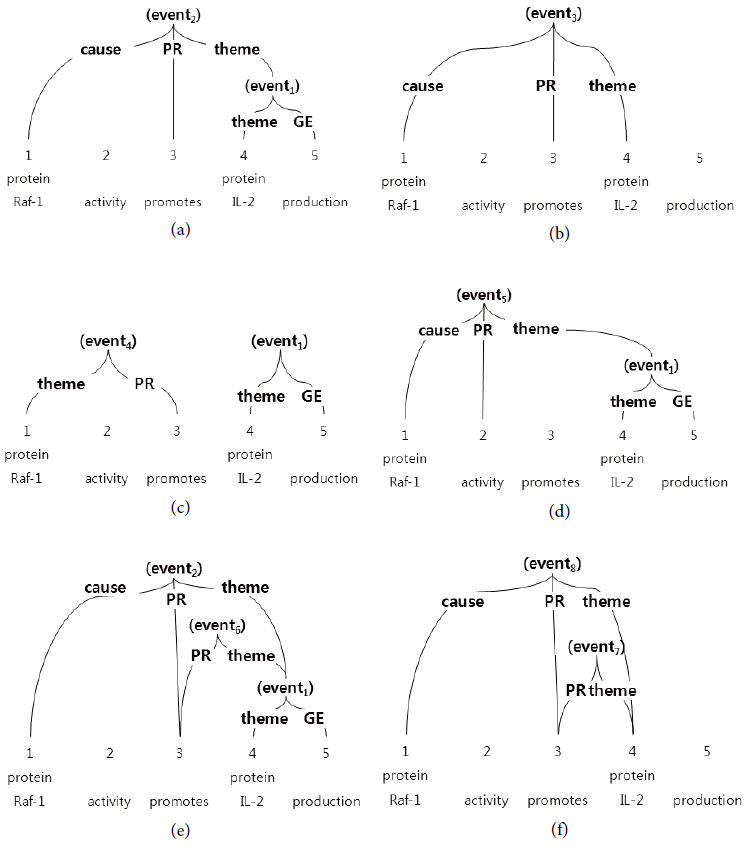
The difficulties of bio-molecular event extraction are shown in Fig. 2. In this figure, only Fig. 2(a) is the correct event extraction from among many other possible candidates. One of the difficult event extractions is the case when an event can take other events as its argument [8,9]. For example, (event2) takes (event1) as its theme argument, as shown in Fig. 2(a). Furthermore, a correct trigger can be missed, such as in Fig. 2(b) and (f) without the gene expression (GE) trigger production. On the opposite hand, an incorrect trigger can be detected, such as in the trigger activity shown in (d). Even if all of the correct triggers are detected, there is the chance that an argument cannot be detected, or that the argument type will be incorrectly identified. Compared with the correct event (event2) at (a), for example, the incorrect event (event4) at (c) takes the incorrect theme argument and no correct cause argument. Even if the correct events are detected, some incorrect events can be unnecessarily detected, as in the event (event6) at (e).
In this paper, we propose a model for bio-molecular event extraction that estimates the probabilities for generating all possible sets of bio-molecular events from a sentence, and that selects the best event set with the highest probability value. The remainder of this paper is organized as follows: Section 2 surveys some previous approaches, and Section 3 explains the proposed model for bio-molecular event extraction. Then, in Section 4, we demonstrate the experimental results, and the characteristics of the proposed model conclude the paper in Section 5.
2. Previous Work
For bio-event extraction, most approaches first detect the triggers in a sentence, and then they obtain the edges that represent the relationship between a trigger and its arguments [7]. Also, they actively utilize dependency parsing information to detect the edges. This is because several previous approaches have already improved their performance by using features extracted from dependency parsing information [4,5,10–12]. Furthermore, the distance between an event trigger and its arguments tends to be much shorter in the dependency path than in the sentence [8]. On the other hand, they can be classified into rule-based approaches, machine learning based approaches, and dictionary and machine learning based approaches.
First, the rule-based approaches automatically draw out some draft event extraction rules from a training set, and then refine these rules that are defined by experts [13–15]. These accurate event extraction rules allow for the rule-based approaches to indicate a comparatively high-precision value. They are very superior to other approaches for simple events. However, these approaches cannot guarantee a reasonable recall on difficult events, including binding and regulations. Additionally, the accuracy can be overly dependent on the expert’s ability. Therefore, modifying the refined rules and changing the features used for constructing the draft rules is a very difficult task.
Second, the machine learning based approaches focus on assigning an event type to an individual token or recognizing an individual relation between a trigger candidate and its argument candidate [8,16,17]. However, the approaches do not mathematically describe how to decompose the problem of extracting the events from a document into the problem of recognizing the individual trigger and the problem of detecting the arguments. Even though these sub problems can use the same machine learning technique with similar features, the approaches also cannot explain the relationship.
Third, the dictionary and machine learning based approaches use a dictionary for the trigger detection, and a machine learning technique for the argument recognition [9,18,19]. However, these approaches are expensive when it comes to building the dictionary, because the dictionary requires the expert’s effort.
In this paper, we propose a model to clearly explain the characteristics of the bio-molecular event extraction problem by using mathematical modeling. In order to clearly describe the connection between the trigger detection step and the argument recognition step, our proposed model changes the event extraction problem into the problem of generating an event table, which includes both unary entries for triggers and binary entries for arguments. For the purpose of significantly simplifying the process of solving an event extraction problem by focusing only on the binary relationship between an event trigger and each of its arguments, the proposed model converts the event table generation problem into the problem of generating each entry in the event table. The proposed model is learned from a training set without using the expert’s support.
3. The Proposed Bio-Molecular Event Extraction Model
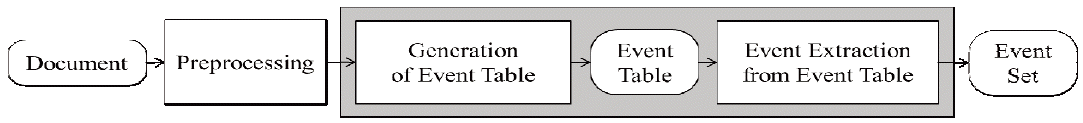
In this chapter, we propose a maximum entropy-based model for bio-molecular event extraction. As shown in Fig. 3, the proposed model consists of a preprocessing step, while analyzing the given document by using natural language processing tools, such as a stemmer, a part-of-speech tagger, and a dependency parser. As the first step in the proposed model, the generation step generates an event table, instead of generating the events themselves. This is done for the purpose of reducing the complexity of solving the event extraction problem, by focusing only on the binary relationship between an event trigger and its argument. Then, the desired events can be easily extracted from the event table. Section 3.1 defines how to estimate the probabilities of generating every entry in the event table. Section 3.3 illustrates the relationship between the event table and a set of the events.
3.1 Generation of the Event Table
The proposed model estimates the probabilities of generating the sets of bio-molecular events from the document DocNE, and selects the best event set E with the highest probability, as represented on the left hand side of Eq. (1). NLP tools, such as a stemmer, a part-of-speech tagger, and a dependency parser, analyze the document DocNE. In order to simplify the event extraction problem, some equations are derived in the following way: first, the document DocNE is divided into two sentence sets of Sφ, which consist of sentences without any named entity; and SNE, which consists of sentences with named entities, as described on the right hand side of Eq. (1).
Because an unnamed entity indicates that there is no event, the sentence set Sφ is removed as presented in Eq. (2). Furthermore, the sentence set SNE and the event set E are replaced with the sentence sequence
S 1 m N E
In order to extract events from an arbitrary sentence by freeing oneself from the position in the sequence
S 1 m N E
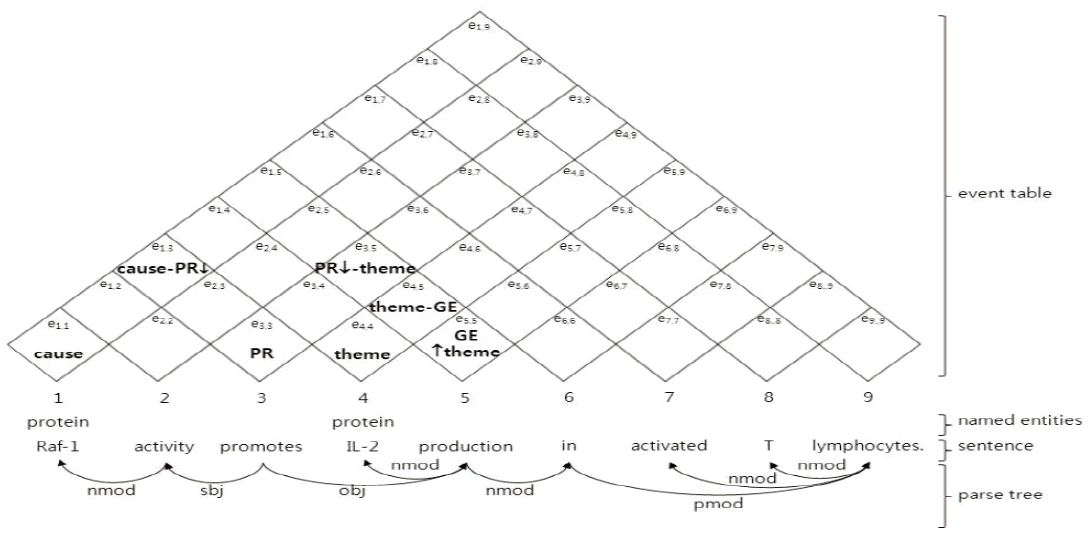
As shown in Fig. 4, the event table consists of entries containing the trigger or argument type. Specifically, entry ex,y represents the trigger-argument relationship between the x-th word wx and the y-th word wy in the sentence. For example, the entry, e4,4 includes the argument type theme, which describes that the fourth word IL-2 will be used for the theme argument of an event. Also, the entry e5,5 contains the type GE↑theme, which represents that the fifth word production will trigger a gene expression event, and then that this gene expression event will be used for the theme argument of another event. In addition, the entry e4,5 includes the type theme-GE, which describes that the word production triggers a complete gene expression event with the theme argument IL-2. Since the symbol ‘↓’ indicates that one event is divided into more than two binary events, the entry e1,3 with cause-PR↓ describes that the entry will be combined with other entries, including the symbol ‘↓’. A more detailed explanation about the event table will be provided in Section 3.3.
3.2 The Maximum Entropy-Based Bio-Molecular Event Extraction Model
In order to solve the event extraction problem by effectively estimating each probabilistic term of Eq. (8), the proposed model utilizes the two words (such as wx and wy); the sentence contexts (such as wx−2, wx−1, wx+1, wx+2, wy−2, wy−1, wy+1, wy+2, and wx+1y−1); the dependency contexts (such as whhx, whx, wdx, wddx, whhy, why, wdy, wddy, and INNERdep(wx,wy)); and the entry histories (such as ehistory), as represented in the equation below.
(10)
(12)
Both Eqs. (11) and (12) provide an example, coupled with a detailed explanation. The word wx includes all of the word itself, its stem, its part-of-speech tag, its form, its named entity tag, and its dependency label, in order to adequately describe the information of the word. The word Raf-1 is represented as Raf-1, Raf-1, noun, Capital: Number, which indicates that the word includes the capital letter R, the number “1,” protein, and noun.
Also, the sentence context consists of the left and right hand side words of each word, and the inner words between the word wx and the word wy in the sentence. Specifically, the context word wi indicates nothing φ if 0 ≥i or i ≥n, where n indicates the number of all words in the sentence. Furthermore, the inner words wx+1y−1 takes nothing φ if x + 1 > y − 1. As shown in Eq. (11), the entry e1,3 can finally utilize w1,w2,w3,w4,w5 as the sentence context.
Additionally, the dependency context is composed of the head and dependent words of each word, and the inner words between the word wx and the word wy in the dependency tree. As presented in the sentence context, wi indicates nothing φ if 0 ≥i, or i ≥n where i substitutes for each of hhx, hx, dx and ddx. As described in Eq. (11), the entry e1,3 can use w1,w2,w3 on the dependency path between the word w1 and the word w3.
Finally, the history context represents some useful entries selected from the event entries previously generated by the chain rule. For example, the entry e1,3 can utilize the immediate event entries e1,1 and e3,3.
In order to practically calculate Eq. (10), the proposed model adopts the maximum entropy framework [22–25], which is one of the most powerful principles of statistical inference. In the maximum entropy framework, the conditional probability of predicting an outcome y given history x is defined as in Eq. (13). In the equation, fi(x, y) is the feature function, and λi is the weighting parameter of fi(x, y). Also, k is the number of features, and Z(x) is the normalization factor for Πy p(x|y)=1. The maximum entropy framework can select a unique joint probability distribution from the set of all joint probability distributions within a reasonable training time [23]. Also, the framework can use arbitrary feature functions in order to reflect the characteristics of the target domain [24]. The ability of freely choosing feature functions gives maximum entropy the obvious advantage over other machine learning methods.
3.3 Relationship between the Event Table and Events
In this section, we first describe the representation of typical events in the event table. And then, we present how the event table can represent the sentences with a distinctive event structure. Every event consists of a trigger and its arguments, where the trigger always indicates a word, while the argument indicates either a word or other event, as shown in Fig. 5(a). Therefore, an event can be represented as a nonterminal node with a few pointers that indicate a trigger or its argument. Also, a trigger and its argument proteins can be represented as terminal nodes without a pointer. Moreover, each event type can be assigned into the event nonterminal node, while either a trigger type or its argument type can be assigned into the terminal node, as is described in Fig. 5(b). In order to reduce the complexity of solving an event extraction problem by focusing on only the event relationship between an event trigger and each of its arguments, the nonterminal is restricted to having two pointers as presented in Fig. 5(c). These terminal and nonterminal nodes can be assigned into the event table (such as the entry ex,x for a terminal node and the entry ex,y with x ≠ y for a nonterminal node). Ultimately, a terminal node for a protein has an argument type, while a terminal node for a trigger has a trigger type. Also, the entry ex,y represents the trigger-argument relationship between the word wx and the word wy in the sentence.
Conversely, three binary events, such as Fig. 5(c), are extracted from the given event table of Fig. 5(d). Then the desired events, such as Fig. 5(a), are obtained by combining two binary events with the symbol ‘↓’ into a single event (such as indicated by the cause- PR-theme in Fig. 5(b)). Specifically, every trigger entry includes as much information as possible, because every trigger leads its event. For example, the trigger type GE↑theme at the event entry e5,5 in the event table Fig. 5(d) denotes that the trigger production will trigger a gene expression event, and that this gene expression event will be used for the theme argument of another event.
Additionally, the proposed event table can cover some unusual examples. As shown in the multiword trigger example of Fig. 6(a), the normal trigger type is assigned to the last trigger word, while the trigger type with B (Begin) or I (Inside) is assigned to other trigger words (shown at the entry e9,9). This is because we assumed that the last word of a trigger leads the trigger. On the other hand, a protein has higher priority than a word in the event table, because a word (such as IFNgamma-induced in Fig. 6(b)) consists of a protein (such as IFNgamma), and a trigger (such as induced). The given correct protein consisting of more than two words (such as the entry e20,20 at Fig. 6(c)) is handled as one element, in order to reduce the event table size without any event extraction performance loss.
As described in the entry e12,12 in Fig. 7(a) and the entry e13,16 in Fig. 7(b), the event table utilizes the symbol “|” to represent more than two triggers or event types [8]. In particular, the entry e13,13 with T |GE indicates that the trigger c-myc can become a transcription event trigger as well as a gene expression event trigger. Also, the entry e13,16 with T-theme and GE-theme indicates that there is both a transcription event and a gene expression event between the same trigger c-myc and the same theme argument protein.
The event table allows for element sharing. The shared element can be a single trigger, such as Fig. 6(c) or Fig. 7(a). Although the trigger induction of Fig. 7(a) is shared by four events, one leads to a gene expression, while the other three lead to positive regulation events. Though both (event1) and (event2) at Fig. 7(b) take the same trigger and the same argument, they can take the different event types, such as e13,16. Thus, part of an event can be shared between two events, such as Fig. 7(c) that (event1) and (event2) take the different theme argument with each other, while they take both the same trigger and the same theme argument.
4. Experiments
In order to examine the practical feasibility of our proposed bio-molecular event extraction model, we evaluated the coverage of the event table and the event extraction performance according to the feature combination. In order to fairly evaluate the proposed model, we utilized the training set, the test set, and the evaluation metrics such as precision, recall, and f-score provided by the BioNLP’09 shared task on event extraction [7].
4.1 Coverage Analysis
For the purpose of examining the coverage of the proposed model, we have applied the correct 8,597 events in the training set to Eqs. (1), (5), and (7), as shown in Table 2, where Num indicates the number of events belonging to each event type. Since Eq. (1) describes the definition of the bio-molecular event extraction problem, there is no coverage loss. Because Eq. (5) cannot extract every event placed in more than two sentences, the coverage of Eq. (5) decreases by 7.99%. Clearly, every trigger and its argument proteins in 529 (6.15%) events are located in different sentences, and 158 (1.84%) events take one of these 529 events as an argument.
Moreover, the coverage of Eq. (7) decreases by an additional 0.65%, because the correct event table does not correspond to the set of the correct events, even though the event table can handle some unusual examples, as previously presented in Section 3.3. For example, the event table cannot include the trigger ‘expression’ in the word ‘overexpression’ without any protein, because the event table is based on a protein unit or a word unit.
4.2 The Effectiveness of Feature Combination
For the purpose of evaluating the bio-molecular event extraction performance according to the feature combination, we utilized some useful features selected from Eq. (10). We also evaluated the proposed model with these features using 10-fold cross validation on the training set. As described in Table 3, the word features indicate wx and wy, which are relative to the type of the entry ex,y in the event table. This feature includes the word itself, its stem, its part-of-speech tag, its form, its named entity tag, and its dependency label, as described in Table 1. Then, the sentence features describe the sentence context of these two words, while the dependency features represent their dependency context. The history features represent some useful entries previously generated by the chain rule.
Table 4 presents a report on the performances of the proposed model on various feature combinations. By adding sentence features or dependency features to word features, the performances tend to increase the recall. Especially, the model adding the sentence features improves the recall by approximately 9% on Regulation events. These results show that the simple word features tend to determine that a given word is a non-trigger word. This is because a trigger word in a sentence frequently occurs as a non-trigger word in other sentences in the training set [8]. However, the model utilizing the sentence features or dependency features comparatively prefer a trigger to an ordinary word based on more precise context information. As the number of correct simple (and binding) events increases, the number of correct regulation events significantly increases by taking these correct simple (and binding) events as arguments. On the other hand, it is remarkable that the sentence features are more useful than the dependency features, because the dependency features can be related to some errors generated by a dependency parser, while the sentence features are free from these errors.
As compared with the model using word features and sentence features, the model adding dependency features or history features improves the precision because of the following reasons since the distance between a trigger and its argument is much closer on the dependency tree than on the raw sentence [8], the model can actually focus on candidate events on the short dependency distance by utilizing the dependency features. In addition, the model can more accurately find the event type by using history features since the history features emphasize the trigger type and the argument type, both of which compose the event type. Finally, Table 4 shows that the model using all kinds of features performs best at a 32.17% f-score because more features generally lead to better performance.
For comparison with previous bio-molecular event extraction models in the same test environment, we evaluated the proposed model on the official evaluation metrics provided by the BioNLP’09 shared task [7]. The proposed model achieved a 50.44% recall, 26.69% precision, and 34.91% f-score in the approximate recursive matching. Compared to the results [7] of other BioNLP’09 shared task participants, the proposed model ranks in the upper-middle range.
5. Conclusion
In this paper, we proposed a maximum entropy-based model for bio-molecular event extraction. The proposed event extraction model has the desirable characteristics, which are described below.
First, we can clearly describe the bio-molecular event extraction problem by mathematical modeling. We mathematically defined the problem to extract the bio-molecular events from a document. We also described a method of deriving the proposed model, which can utilize many useful features, such as sentence contexts and dependency contexts, from the definition.
Second, we can claim that it is possible to replace the bio-molecular event extraction problem with the event table generation problem. In the proposed event table, each unary entry is assigned to each word in the sentence, and each binary entry represents the trigger-argument relationship between the given pair of words. The event table can cover some unusual events, such as the multi-word trigger or element sharing, as described in Figs. 6 and 7. Our experimental results showed that the coverage of bio-molecular event extraction decreases only by 0.65%. This is because the correct event table does not correspond to the set of the correct events.
Third, we can simplify the process of solving a bio-molecular event extraction problem because the proposed model focuses only on the individual binary relationship between an event trigger and each of its arguments. Specifically, we replaced the event extraction problem with the event table generation problem, and then we decomposed the event table generation problem into the small manageable problems of filling each entry into the event table.
Fourth, we verified the useful feature combination by analyzing the event extraction power of each feature combination. We have found that more features mainly tend to significantly increasing the precision of the bio-molecular event extraction problem, even though more features slightly decreased the recall in some cases. Furthermore, it is remarkable that the sentence features are more useful than the dependency features, since the dependency features can be related to some errors generated by a dependency parser, while the sentence features are free from these errors.
Unlike previous approaches that have intuitively designed a pipeline consisting of a trigger detection step and an argument recognition step, the proposed model combines these two steps by using the event table. Also, the model is designed based on the mathematical derivation process rather than on intuition. Furthermore, the proposed model can describe why these two steps can utilize the same machine learning techniques and similar features.
For future research, we would like to study a method of selecting more appropriate features for the maximum entropy model, in order to extract the events more precisely. In addition, we aim at applying an anaphora resolution method to the bio-molecular event extraction problem for covering some events in which a trigger and arguments are located in different sentences.